Raising Young Cuckoos
It is not appropriate to conduct multiple comparisons in this example because the H0 in the one-way ANOVA was not rejected. This result suggests that there is no difference among group means and, thus, no need to determine which specific groups differ.
Temperature and Turtle Hatchings
- On the first question, define any symbols used in the hypotheses.
- On the second question, use the p-value from the ANOVA table. That is the p-value that assesses whether all group means are equal or not. Tukey’s multiple comparisons are only used to assess difference in paired means AFTER it has been determined that there is a difference in means.
- On teh second and third questions, make sure to include the p-value (see below). The p-value is the evidence for your statement … don’t just make a statement without showing your evidence. Also, I would like to see the p-value in your sentence so it is clear to me what you are looking at … don’t just say “as seen below” (because there is lots of information below).
- In the fourth question, clearly indicate which group is greater (or lesser). Don’t just say that you are 95% confident that the difference is between such-and-such.
- In the fifth question include a plot of Tukey’s corrected means with confidence intervals.
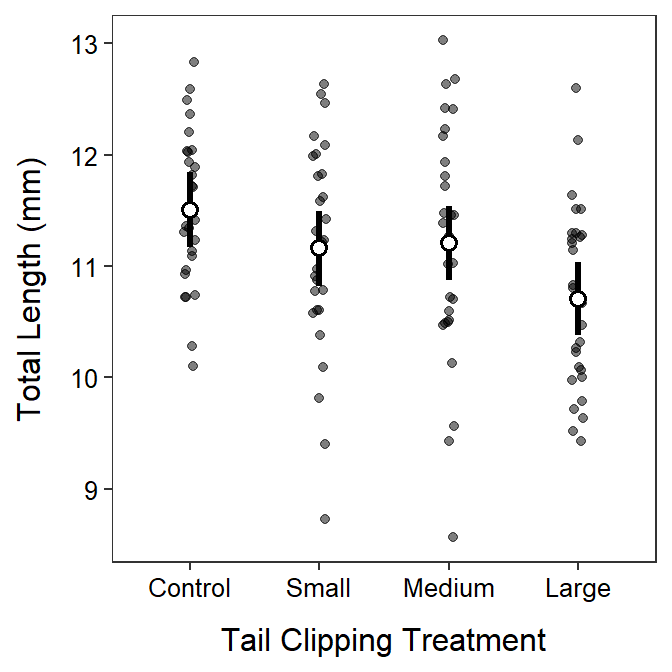
- In the last question, note that there is no difference in means for the first three temperatures. So, even though, the plot shows slight differences in the SAMPLE means, those differences were not great enough to conclude that the POPULATION means were different. So, your conclusion should not make it sound like there is a difference among those temperature treatments.
- Also note in the fourth, fifth, and last questions that the conclusions are about the MEAN days to hatch not days to hatch because that is what is tested in the hypotheses. Make sure to include the word MEAN.
- Note how concise the answer key is. Please work to get your answers this concise.
- The H0: μ15C=μ20C=μ25C=μ30C, where μ is the mean days to hatch and the subscripts are the temperatures in the temperature treatments. The HA: “mean days to hatch differs between at least one pair of treatment temperatures”.
- It appears that the mean days to hatch differs between at least one pair of treatment temperatures (p<0.00005<α).
- It appears that the mean days to hatch differs between 30oC and 15oC (p<0.00005), 20oC (p=0.0008), and 25oC (p=0.0001). The mean days to hatch did not differ between any other pairs of temperature treatments (p>0.0983).
- It appears that the mean days to hatch at 30oC was between 22.7 and 53.9 days lower than at 15oC, between 8.9 and 40.1 days lower than at 20oC, and between 13.5 and 44.7 days lower than at 25oC.
- The plot of means with 95% confidence intervals is shown below.
- It appears that temperature does not affect the mean days to hatch from between 15oC to 25oC, but declines dramatically from 25oC to 30oC. Thus, temperature does affect the mean days to hatch but not until temperature is above 25oC.
R Code and Results.
> d <- read.csv("turtles.csv")
> d$Temperature <- factor(d$Temperature)> lm1 <- lm(Days~Temperature,data=d)
> anova(lm1)Analysis of Variance Table
Response: Days
Df Sum Sq Mean Sq F value Pr(>F)
Temperature 3 8025.5 2675.16 15.978 9.082e-07
Residuals 36 6027.3 167.42 > mc <- emmeans(lm1,specs=pairwise~Temperature)
> ( mcsum <- summary(mc,infer=TRUE) )$emmeans
Temperature emmean SE df lower.CL upper.CL t.ratio p.value
15 58.4 4.09 36 50.1 66.7 14.273 <.0001
20 44.6 4.09 36 36.3 52.9 10.900 <.0001
25 49.2 4.09 36 40.9 57.5 12.024 <.0001
30 20.1 4.09 36 11.8 28.4 4.912 <.0001
Confidence level used: 0.95
$contrasts
contrast estimate SE df lower.CL upper.CL t.ratio p.value
15 - 20 13.8 5.79 36 -1.78 29.4 2.385 0.0983
15 - 25 9.2 5.79 36 -6.38 24.8 1.590 0.3970
15 - 30 38.3 5.79 36 22.72 53.9 6.619 <.0001
20 - 25 -4.6 5.79 36 -20.18 11.0 -0.795 0.8563
20 - 30 24.5 5.79 36 8.92 40.1 4.234 0.0008
25 - 30 29.1 5.79 36 13.52 44.7 5.029 0.0001
Confidence level used: 0.95
Conf-level adjustment: tukey method for comparing a family of 4 estimates
P value adjustment: tukey method for comparing a family of 4 estimates > ggplot(data=d,mapping=aes(x=Temperature,y=Days)) +
geom_jitter(alpha=0.5,width=0.05) +
geom_pointrange(data=mcsum$emmeans,
mapping=aes(x=Temperature,y=emmean,
ymin=lower.CL,ymax=upper.CL),
size=1.1,fatten=2,pch=21,fill="white") +
labs(y="Days to Hatch",x="Temperature Treatment") +
theme_NCStats()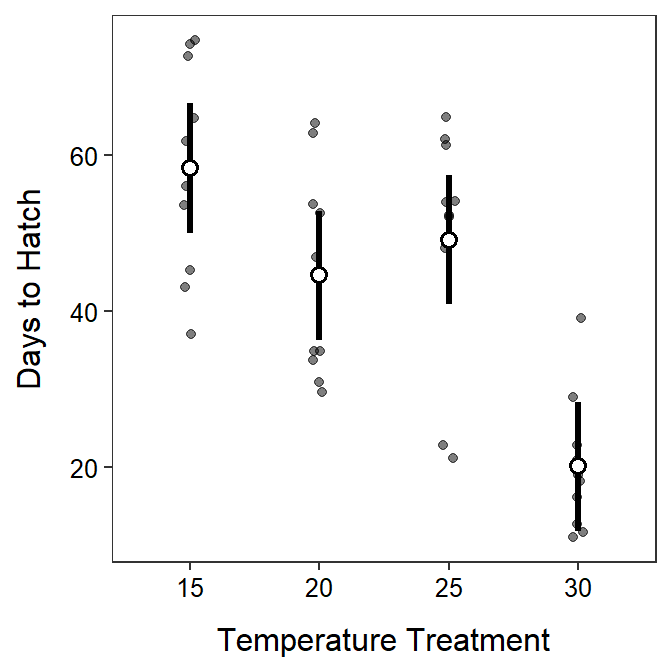
Salamander
- The H0: μControl=μSmall=μMedium=μLarge, where μ is the mean total length at the end of the experiment and the subscripts are the tail clipping treatments. The HA: “mean total length differs between at least one pair of tail clipping treatments”.
- It appears that the total length differs between at least one pair of tail clipping treatments (p=0.0102<α).
- It appears that the mean total length differs only between the control (no clipping) treatment and the “large” (10.0-mm) clipping treatment (p=0.0051). The mean total length did not differ between any other pairs of tail clipping treatments (p>0.1512).
- It appears that the mean total length was between 0.2 and 1.4 mm lower in the “large” clipping treatment than the control (no clipping) treatment.
- The plot of means with 95% confidence intervals is shown below.
- It appears that clipping only results in a difference in mean total length if the large (10.0-mm) clipping is compared to no clipping (i.e., control). Part of the reason that few differences in mean total length were observed is because there is a large amount of natural variability among individuals in each group.
R Code and Results.
df <- read.csv("https://raw.githubusercontent.com/droglenc/NCData/master/Salamanders.csv")
df$Treatment <- factor(df$Treatment,levels=c("Control","Small","Medium","Large"))
lm2 <- lm(TL~Treatment,data=df)
anova(lm2)Analysis of Variance Table
Response: TL
Df Sum Sq Mean Sq F value Pr(>F)
Treatment 3 9.348 3.1161 3.953 0.01017
Residuals 109 85.924 0.7883 mcS <- emmeans(lm2,specs=pairwise~Treatment)
( mcSsum <- summary(mcS,infer=TRUE) )$emmeans
Treatment emmean SE df lower.CL upper.CL t.ratio p.value
Control 11.5 0.168 109 11.2 11.8 68.581 <.0001
Small 11.2 0.168 109 10.8 11.5 66.495 <.0001
Medium 11.2 0.168 109 10.9 11.5 66.793 <.0001
Large 10.7 0.165 109 10.4 11.0 64.941 <.0001
Confidence level used: 0.95
$contrasts
contrast estimate SE df lower.CL upper.CL t.ratio p.value
Control - Small 0.35 0.237 109 -0.269 0.969 1.475 0.4561
Control - Medium 0.30 0.237 109 -0.319 0.919 1.264 0.5875
Control - Large 0.80 0.235 109 0.186 1.414 3.402 0.0051
Small - Medium -0.05 0.237 109 -0.669 0.569 -0.211 0.9967
Small - Large 0.45 0.235 109 -0.164 1.064 1.914 0.2282
Medium - Large 0.50 0.235 109 -0.114 1.114 2.127 0.1512
Confidence level used: 0.95
Conf-level adjustment: tukey method for comparing a family of 4 estimates
P value adjustment: tukey method for comparing a family of 4 estimates ggplot(data=df,mapping=aes(x=Treatment,y=TL)) +
geom_jitter(alpha=0.5,width=0.05) +
geom_pointrange(data=mcSsum$emmeans,
mapping=aes(x=Treatment,y=emmean,ymin=lower.CL,ymax=upper.CL),
size=1.1,fatten=2,pch=21,fill="white") +
labs(y="Total Length (mm)",x="Tail Clipping Treatment") +
theme_NCStats()